Software and digital applications - Sustainable agriculture
- Home
- Work with us
- Cirad'Innov solutions
- Products and services
- Gigwa
Gigwa: High-throughput genotyping data management system
Giwa © Cirad
Simplified analysis of large volumes of genomic data
How can we better assess and use the biodiversity of plants cultivated in tropical countries? This ambitious goal was the aim of the European ARCAD project, from which the Gigwa solution emerged. Born of CIRAD's and IRD's expertise in bioinformatics, the system was initially designed to facilitate the centralization and handling of the large volumes of genotypic data generated by the emergence of NGS sequencing. Its latest adaptations now allow this data to be shared, visualized and analyzed. This tool is constantly evolving to serve the widest possible range of applications in both agronomy and health sectors.
Featuring an ergonomic web interface, capable of adapting to animal, human or plant data, and handling both the metadata associated with biological material and the functional annotations of variants, Gigwa is a solution that lends itself to a wide range of uses. It can be used as a data warehouse, as a converter between different formats, as an online filtering, visualization and analysis tool, and as a source of interoperable data for workflows of all kinds.
With the support of our development team, the system can be integrated with other web platforms (e.g. Genome-Hubs, PeanutBase, Musa Germplasm Information System, Breeding Management System), and additional functionalities can be developed on request.
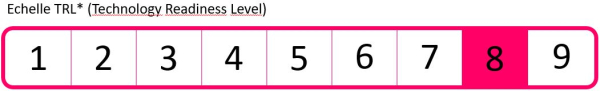
Stage of development
TRL8 - Qualification of the complete system in an operational environment
The benefits of the Gigwa solution
Gigwa, Genotype Investigator for Genome-Wide Analysis, offers a wide range of services. The platform :
- is able to manage up to tens of billions of genotypes;
- can be easily installed on a local workstation or on a server, using a Docker image;
- can be deployed in multi-user mode, managing single sign-on (SSO) for easy institutional use;
- is interoperable via various REST API standards (BrAPI v1 & V2, GA4GH);
- supports a wide range of data import and export formats;
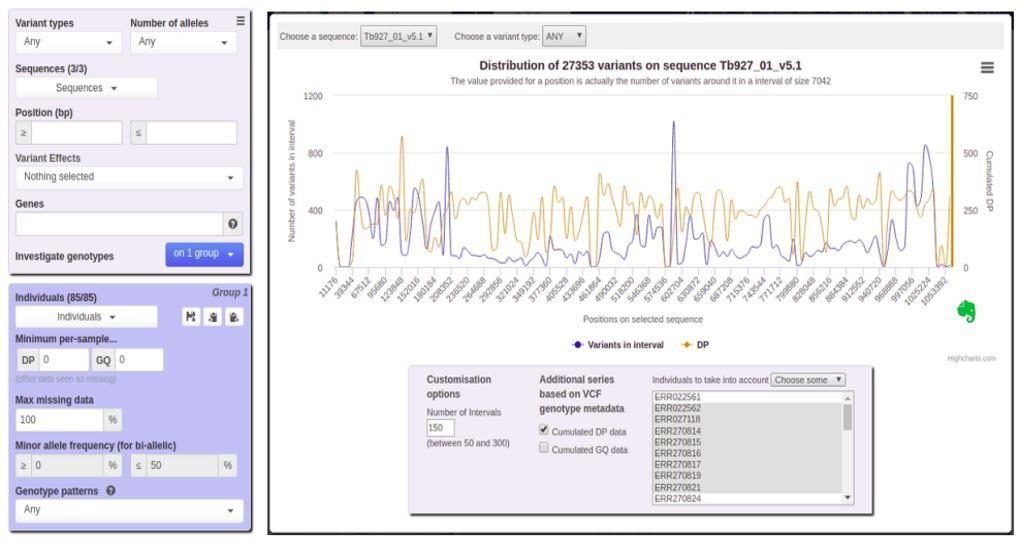
- offers filtering of variant characteristics (position, annotations, etc.), based on polymorphism between individuals or on data quality criteria;
- accepts different levels of ploidy and phased data;
- can archive search criteria in the form of favourites;
- features a search function for discriminating variants between 2 groups of individuals ;
- allows graphical representation of statistics calculated in real time (distribution of variants, MAF, Fst, Tajima's D);
- includes integrated visualization and analysis tools (IGV.js, Flapjack-Bytes);
- facilitates connection with external tools for further analysis (e.g. Galaxy, SniPlay, etc.);
- includes a database backup and restore functionality for enhanced security to safeguard critical data from loss.
A tool to support those involved in sequencing data analysis
At the service of genomics applied to varietal or animal selection and diagnosis
Gigwa's functionalities can meet a wide range of needs for companies involved in genomic data analysis. The tool is quite intuitive, enabling researchers to get to grips with it without necessarily calling on the services of a bioinformatician. Compared with other free open source tools on the market, it offers a number of complementary applications, including the ability to compare 2 groups of individuals on the basis of criteria of interest.
The openness of the system (open source code and interoperability through APIs) is also of interest to companies with an in-house solution, for which Gigwa can provide additional functionality and serve as a platform for the development of modules to meet specific needs.
Serving genetics and genomics education
Many open-source tools exist, but they don't offer Gigwa's power or ease of use. With over 40 years' expertise in the genomic study of tropical and Mediterranean plants, we are keen to support training and teaching establishments with useful, high-performance tools. Gigwa is currently being used as a working tool for case studies submitted to Master 2 students at the Universities of Montpellier, Dakar and Antananarivo as part of a module taught by CIRAD researchers and engineers.
How can I obtain Gigwa services?
An instance of Gigwa hosted by CIRAD is freely available at https://gigwa.southgreen.fr/gigwa/
If you would like to see how the tool works, 4 demonstration videos are available:
- Import your data onto the platform;
- Filter your data;
- Make the most of your filtered data;
- Discriminate variants according to phenotypes of interest.
Are you interested in Gigwa and would like to explore the possibility of using the tool to manage your own genomic variation data, integrate it into your information system or develop additional modules? Contact us: ciradinnov@cirad.fr
The research teams
The community of developers and regular users contributing to the evolution of the Gigwa system.
This tool was made possible through the collaboration of three local units. :
INTERTRIP: https://umr-intertryp.cirad.fr/en
AGAP: https://umr-agap.cirad.fr/en
DIADE :https://www.cirad.fr/en/about-us/research-units/diversity-adaptation-development-of-plants
References and intellectual property
Source code
Fully available under the GNU Affero General Public License v3.0 at the following address https://github.com/SouthGreenPlatform/Gigwa2
Registered with the Agence Protectrice des Programmes on 05/04/2019 under number IDDN1 .FR2 .0013 .1500364 .0005 .S6 .C7 .20198 .0009 .2090010
Publications
Managing high-density genotyping data with Gigwa. Sempere Guilhem, Larmande Pierre, Rouard Mathieu. 2022. In : Plant Bioinformatics: methods and protocols. Edwards David (ed.), 415-427. (Methods in Molecular Biology, 2443) ISBN 978-1-0716-2067-0 https://doi.org/10.1007/978-1-0716-2067-0_21
Gigwa v2—Extended and improved genotype investigator. Sempere Guilhem, Petel Adrien, Rouard Mathieu, Frouin Julien, Hueber Yann, De Bellis Fabien, Larmande Pierre. 2019. GigaScience, 8 (5), 9 p. https://doi.org/10.1093/gigascience/giz051
Gigwa—Genotype investigator for genome-wide analyses. Sempere Guilhem, Philippe Florian, Dereeper Alexis, Ruiz Manuel, Sarah Gautier, Larmande Pierre. 2016. GigaScience, 5 (25), 9 p. https://doi.org/10.1186/s13742-016-0131-8